TUTORIAL
ODCs (Orphan Disease Connections) is a web-based application to explore potential relationships between rare diseases established
through the integration of disease susceptibility genes and protein-protein interactions.
The current version contains 54,941 relationships between 3,032 diseases and 3,061 genes.
See all the rare diseases and genes included in ODCs
here.
HOW TO USE ODCs?
Interested in one disease and its potential connections? Click on
Rare disease search.
Interested in exploring the connections of two diseases? Click on
Diseases connection search.
Interested in one gene or protein? Click on
Gene search.
THE DATA
ODCs integrates two main sources of data:
Additionally, ODCs provides links out to several resources:
-
ICD-10 (international classification of diseases),
-
Orphanet (rare disease and orphan drug database),
-
OMIM (catalog of human genes and genetic disorders),
-
UniProt (protein sequence and functional information),
-
MeSH (medical controlled vocabulary).
As well as facilitating searches to the
PubMed.
RARE DISEASE SEARCH
Introduce a valid rare disease name or synonym and click the "Search" button to display information on that disease as well as its potential relationships
with other rare diseases.
For example, this is the output provided for
Rett syndrome.
A summary panel on the right displays OMIM references, the number of associated genes and signs/symptoms (from Orphadata),
as well as the number of connected diseases calculated by OCDs. The details are shown on the tabs below the panel.
Connected diseases are also shown graphically as a network on the left:
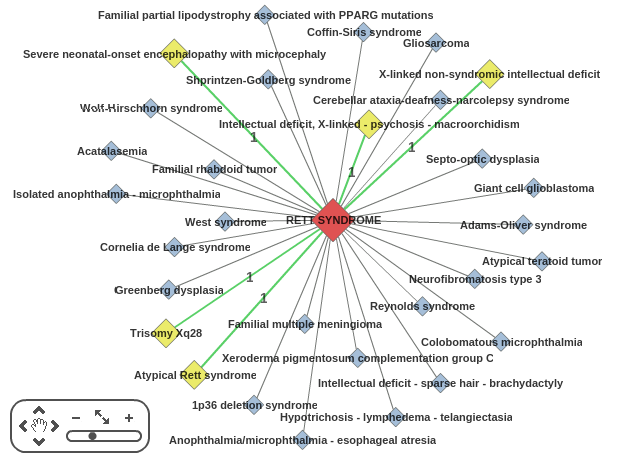
The central red node is the disease of interest (in this case, Rett syndrome).
Yellow nodes represent diseases that share at least one susceptibility gene with the central disease (bigger nodes mean more susceptibility genes shared).
Blue nodes represent diseases that are connected through protein-protein interactions.
A panel in the bottom left corner allows the user to zoom the network diagram.
Click on a yellow or blue node. This will show two additional buttons at the bottom of the graph:
the first one links out to the corresponding disease page in Orphanet; the second, displays
the details of the connection between the disease selected and Rett syndrome in ODCs (as if you perform a
Diseases connection search).
You can export the network data clicking on the button at the top of the graph (CSV file).
DISEASES CONNECTION SEARCH
Introduce two valid rare disease names or synonyms and click the "Search" button to explore their potential relationship. ODCs establishes
relationships through disease susceptibility genes and corresponding protein-protein interactions.
As an example you can search
Noonan syndrome and Costello syndrome.
The right panel shows the summary information on each disease, as well as the summary relationships (in this
case one common gene, 5 protein-protein interactions and 13 common signs/symptoms).
The details can be seen on the graphical display on the left:
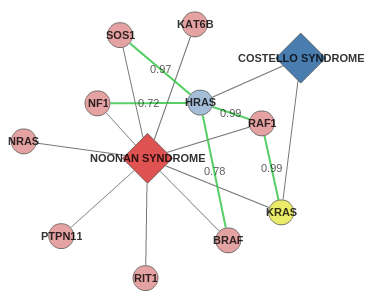
Gene/protein nodes are represented as circles, colored by the same color as their corresponding disease. Yellow circles show
genes shared by the two diseases.
In this example both Noonan and Costello syndromes share one gene, KRAS, and their relationship can
also be explained by the interactions between HRAS and RAF1, SOS1, BRAF and NF1. The scores of each
protein-protein interaction in HIPPIE are also displayed in the graph.
You can also select one gene/protein node to access information in UniProt (click the UniProt button)
or to explore other disease associations, as if you perform a
Gene search (click the ODCs button).
Click on the "Export data" button to download the information shown in the graph.
In the previous example the connection between Noonan and Costello syndromes was supported both by a common gene, and
a set of protein-protein interactions. There are other associations that can only be established on protein-protein interactions,
like the
following example:
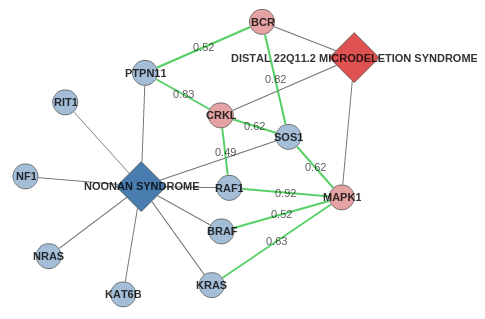
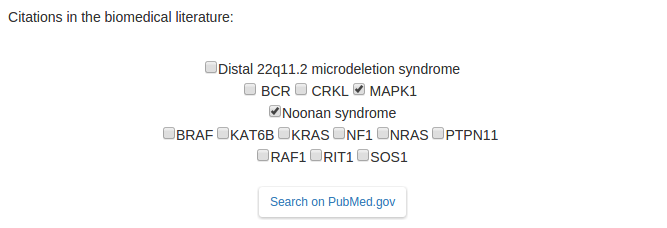
Explore the tabs to see the diseases genes (links out to UniProt and OMIM) and interactome associations (links out to evidences on HIPPIE). Now
you can easily search the biomedical literature (click on
PubMed search), select disease names and/or genes (e.g.
Noonan syndrome and MAPK1), and finally click on the "Search on PubMed.gov" button.
 GENE SEARCH
GENE SEARCH
In this search, ODCs shows the rare diseases associated with a gene. Introduce a valid gene name,
for example
SOX10, and click the "Search" button.
A summary panel on the right shows that SOX10 is associated with 4 rare disases and has been described to interact
with 11 proteins. External references (UniProt and OMIM) are also available. More details on the tabs below the panel.
A graph displays in a network this information:
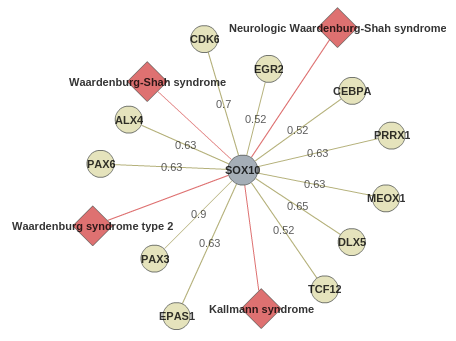
Genes/proteins are represented as circles, centered around SOX10 in grey.
HIPPIE scores are shown for each protein-protein interaction.
Associated diseases are shown as red diamonds.
You can click on any gene/protein to access further information in UniProt or to display a
new ODCs search centered on that gene.
Click on any rare disease to access further information in Orphanet or to display a
new ODCs search centered on that disease.
An "Export data" button is also available.
ADDITIONAL INFORMATION
ODCs is optimized for PCs and mobile devices. For a correct visualization of the networks a modern browser with the Flash plugin installed is needed.
The DISEASE SEARCH and the DISEASE CONNECTION SEARCH using Orpha number from Orphanet is also available:
http://csbg.cnb.csic.es/odcs/disease_showresults.php?orphanum0=778 (Rett syndrome)
http://csbg.cnb.csic.es/odcs/connection_showresults.php?orphanum1=648&orphanum2=3071 (Noonan and Costello syndromes).
-
| 2014 |
Computational Systems Biology Group |
Spanish National Center for Biotechnology (CNB-CSIC)
|
CSIC |
Escuela Politécnica Superior |
UAM | Designed & developed by Sara Fernández Novo |




