|
Sequence Analysis and Structure Prediction Service at the National Center of Biotechnology
(CNB-CSIC)
Relevance of sequence analysis in Molecular Biology
 Sequence analysis and protein structure prediction have been shown to be very useful tools for Molecular Biology. These approaches, that can be applied in a wide-ranging way to multiple areas, well used can restrict experimental work. Starting from the primary sequence of a gene, using these tools you could obtain data on evolution, functional/structural domain organization, functionally relevant regions or residues, functional hypothesis, possible subcellular localisation, interactions with other genes, structural models, etc...(published examples). Many of these tools can be easily used by the community through web interfaces, while others are either harder to use or the results are difficult to interpret without previous bioinformatic knowledge. Also, many bioinformatic protocols are not easy to implement in automatic tools and they need manual intervention or expertise to be carried out.
Sequence analysis and protein structure prediction have been shown to be very useful tools for Molecular Biology. These approaches, that can be applied in a wide-ranging way to multiple areas, well used can restrict experimental work. Starting from the primary sequence of a gene, using these tools you could obtain data on evolution, functional/structural domain organization, functionally relevant regions or residues, functional hypothesis, possible subcellular localisation, interactions with other genes, structural models, etc...(published examples). Many of these tools can be easily used by the community through web interfaces, while others are either harder to use or the results are difficult to interpret without previous bioinformatic knowledge. Also, many bioinformatic protocols are not easy to implement in automatic tools and they need manual intervention or expertise to be carried out.
Service Functions
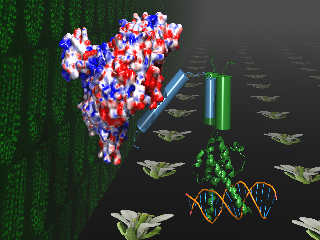 The main function of the service is to give bioinformatic support in sequence analysis and structure prediction to all groups at the CNB that could require it. This includes, among other things:
The main function of the service is to give bioinformatic support in sequence analysis and structure prediction to all groups at the CNB that could require it. This includes, among other things:
Localization of homologous gene within different genomes, totally or partially sequenced, using high sensibility methods.
Generation of high quality multiple sequence alignment.
Evolution: gene distribution within organisms/kingdoms, phylogenetic tree generation,...
Subcellular localization prediction
Structural/functional domains localization
Prediction of functionally relevant residues or regions
Prediction of post-translational modification sites (phosphorylation, glycosylation,..)
Prediction of interaction sites.
Prediction of functional relationships with other genes by cotext methods(not based on homology) - Prediction of physical/functional interactions
Generation of structural organisation drafts: globular domain localization, unstructured regions, coiled-coil, transmembrane regions, etc...
Secondary structure prediction
3D structure prediction for close and remote homology
Generation of high-quality structural representations for publications
Additionally, the service will organize in collaboration with others, periodic courses and seminars about the use and possibilities of the bioinformatic tools available.
Besides, as far as possible, we would like to continue the recently initiated line of "a-la-carte" software development (bespoke solutions) for experimental groups. The idea is to implement, using modern and efficient technologies, protocols that now these groups are either carrying out with general tools (i.e. Excel, FileMaker, Access, etc...) or simply not carrying out because of the impossibility of doing it with the available software.
Working model
|
The sequence analysis and structure prediction services are offered under a "collaboration" model. This is to say, without charging and signing the papers that could eventually emerge from that collaboration, only if the work done by the service is worthy. For the particular case of "a-la-carte" software development, this will be done only if the developed application has enough general interest to be published.
|
Staff
The following people will be part-time dedicate to this service:
Florencio Pazos Cabaleiro
Monica Chagoyen Quiles
Juan Carlos Sanchez Ferrero
Contact
Florencio Pazos
Lab. B-18
Tlf. 91.5854669
|
Related links
On line tools for sequence analysis (Expasy)
Bioinformatics Tools @ EBI
Automatic sequence analysis tools @ CNB
|

