Figure 1
The CoMentG server allows to interactively browse the relationships between biomedical entities inferred from co-mentions in the scientific literature. The procedure for inferring these relationships is described in detail in:
Please cite that reference when reporting any data or result obtained from this server.
The access to this server is free to everyone.
This tutorial is intended to make you familiar with the main features of the server. The contents of the tutorial are based on the sets of relationships between nine sets of biomedical concepts available at https://csbg.cnb.csic.es/comentG/. You can open that instance of the CoMentG server in a new browser window/tab in order to follow this tutorial. These 9 datasets include tens of thousands of biomedical concepts related to diseases (DOID and MONDO ontologies), biological processes, molecular functions and cell compartments (GO), clinical signs (HPO), cell types (CL ontology), tissues and body parts (UBERON), bacteria (MeSH terms related to microorganisms) and chemical compounds (human-related metabolites from HMDB and MeSH terms related to chemical compounds). Take a look at the help section of the link above for more information on these datasets and their co-mention relationships.
In this tutorial, we are going to look for comentions between Alzheimer's disease and clinical signs, biological processes/molecular functions, cell types and chemical compounds, in order to have a picture of the different molecular aspects of this disease.
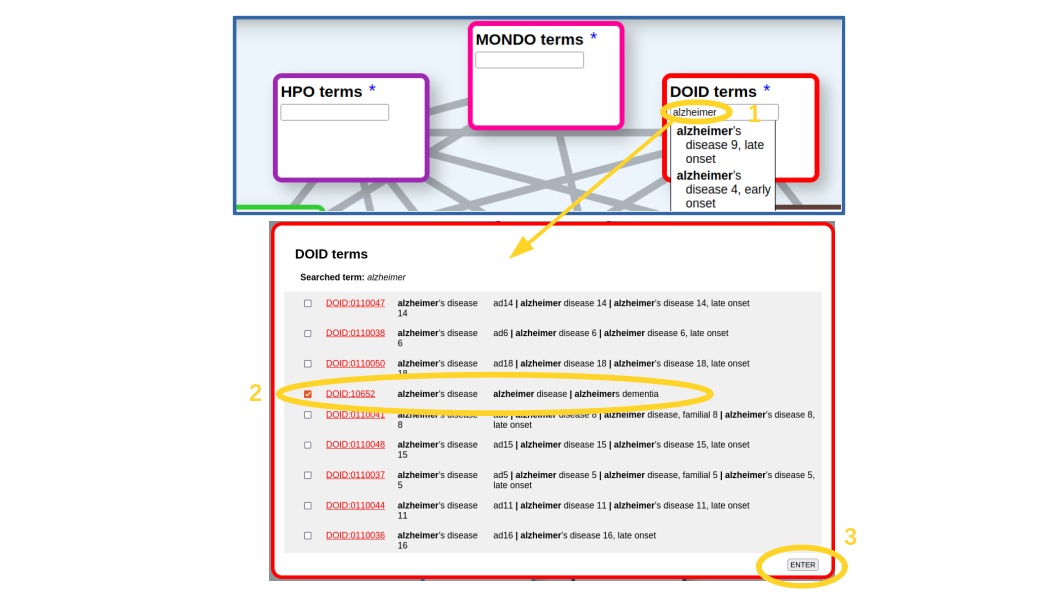
The first step is to look for our term(s) of interest in the corresponding ontology. In this case, we are going to look for the term corresponding to "alzheimer's disease" on the "disease ontology" (DOID). For that we start typing the term in the corresponding textbox (Figure 1). An autocompletion list shows up. It includes all terms with our typed characters in the name or any of the synonyms (what in this case are mainly variants of this disease). If our term of interest is on the list, we just select it, if not, we press [Enter] and a more detailed list of the matching terms pops up (Figure 1), which includes the terms' IDs (hyperlinked to the corresponding entries in the original resource to eventually retrieve more info), their names and synonyms. In this list we can select one or more terms (in case we want, for example, to retrieve relationships for more than one Alzheimer variants). In our case, we select only the main entry for the disease (DOID:10652), as we are not interested in an specific variant, and press the [Enter] button (Figure 1).

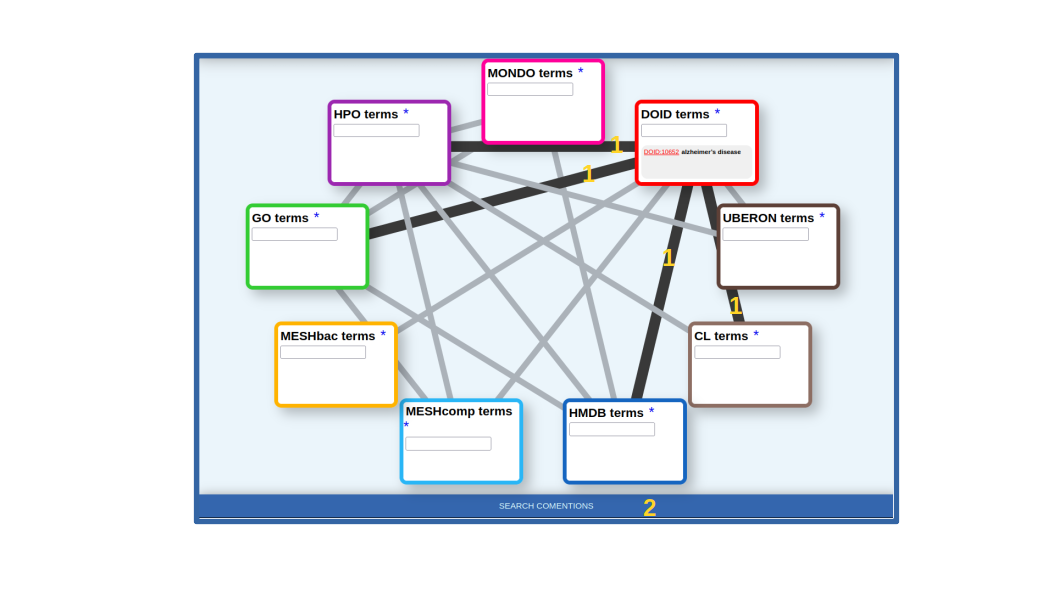
Once the term or terms of interest are in the corresponding box, we select the lines connecting it with all the other datasets for which we want to retrieve relationships. In our case, the lines connecting DOID with HPO (symptoms, clinical signs), GO (gene functions), HMDB (human-related metabolites) and CL (cell types) (Figure 2). Once the term(s) and the desired types of relationships are choosen, we press the [SEARCH COMENTIONS] button (Figure 2).
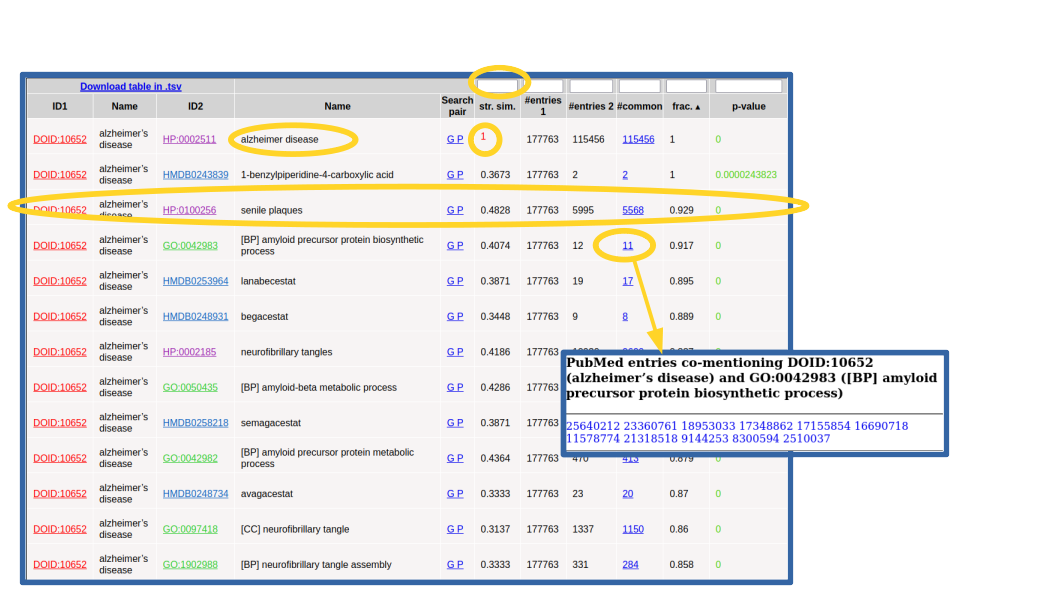
The co-mention relationships between the term(s) of interest and those terms in the other selected resources are shown in a table. The top of this list for our example is shown in Figure 3.
For each relationship in the list (pair of terms), the IDs and names of both terms are shown, being the IDs links to the entries for these terms in the corresponding resources. There are two links for performing simple text searches of both terms together in Google (“G”) and PubMed (“P”) to further dig into the relationship. These searches do not include synonyms but just the names shown in the table. The “string similarity” between both terms (including all their synonyms, not shown in this list) is also indicated, and cases of high similarity (≥0.7) are highlighted in red color. This is to highlight trivial co-mentions due to identical or very similar terms.
The next columns show the number of PubMed entries mentioning the first term, those mentioning the second, those mentioning both together, and the ratio of that respect to the minimum of the first two. By default, the list is sorted by this parameter ("frac"). If you click in the number of articles mentioning both terms together you get a list with the PubMed IDs of these articles, which are links to the corresponding papers (Figure 3). Note that this set of papers can be different from those you get with the “P” link above due to various reasons, but mainly to the fact that the “P” link does not include synonyms.
The final column shows the p-value of the hypergeometric test used for assessing the significance of the co-mention (see the publication above). Only pairs with p-value ≤ 0.001 are shown, and those with p-value ≤ 1·10-5 are highlighted in green color. The table can be sorted by any column by clicking the corresponding header. At the top left corner of the table there is a link to download the whole table in .TSV format so that it can be further processed, for example importing it into an external spreadsheet program. There are also text-boxes above the headers of the numerical columns to restrict the list of relationships according with certain values: less or equal (<=) the entered value for "str. sim.", >= for "#entries", >= for "frac" and <= for "p-value".
For example, for the relationship between "Alzheimer disease" and its well-known symptom "senile plaques" (Figure 3), 177,763 PubMed entries mention the disease, 5,995 mention the symptom ("senile plaques"), and most of these (5,568 (92.9%)) mention both terms together, yielding a p-value of 0.0.
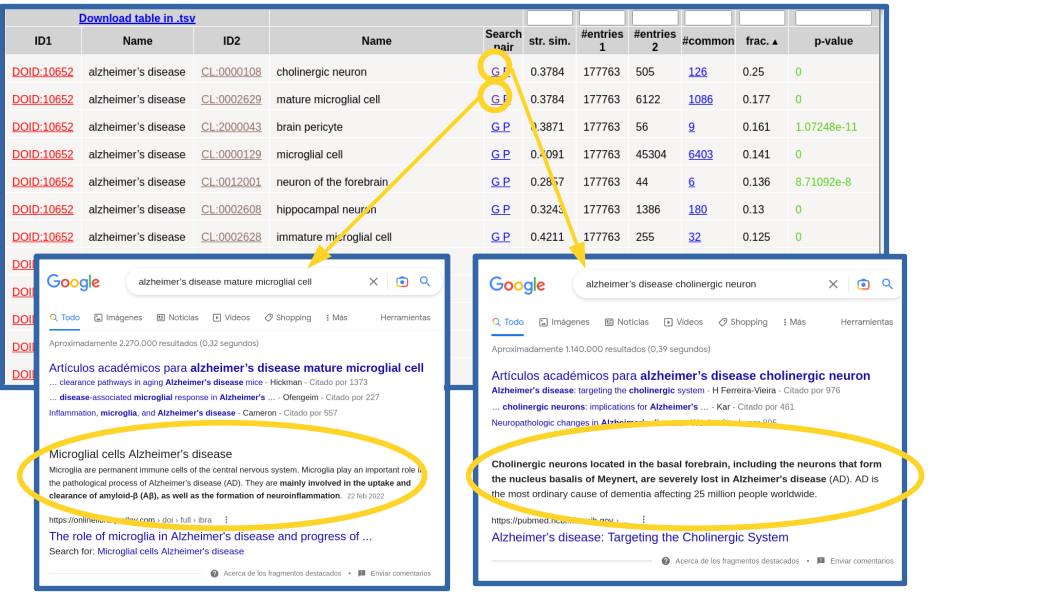
In this example, all the terms that show up in the list make sense considering what we intuitively known about this complex pathology and, together, they provide a comprehensive picture of its internal causes (molecular processes), external symptoms, cell types it affects, and related drugs and chemical compounds (Figure 3). Regarding clinical signs (HPO), we see the typical molecular/physiological manifestations related to senile plaques, neurofibrillary tangles, cortical atrophy, hirano bodies, etc, as well as its higher level mental and behavioral symptoms (apathy, delusions, dementia, memory problems, …). Within the biological processes terms (GO[BP]), we find those related to β-amyloid metabolism and neurofibrillary tangles, among others. Regarding molecular functions (GO[MF]), we can see those related to binding to amyloid-beta and tau protein, among others. Also the “cellular compartments” category of GO (GO[CC]) reflects what is known on this disease at the level of sub-cellular structures, with terms such as gamma-secretase complex, alpha-ketoglutarate dehydrogenase complex, lewy bodies, etc. Regarding the cell types (CL), we see the brain-related cell types associated to this disease, being the first ones "cholinergic neuron" (CL:0000108), known to be severely affected in this disease, and "mature microglial cell" (CL:0002629), which are involved in the uptake and clearance of β-amyloid. In Figure 4 we use the "G" link to perform Google searches for these cell type relationships in order to dig into their involvemetn in Alzheimer. For the chemical compounds (HMDB), the found relationships are mainly with drugs used for treating this disease, such as lanabecestat, begacestat, semagacestat, etc. If you want to inspect one of this categories in detail it is better to restrict the search for relationships to it in the previous step (Figure 2 above), as done for cell types (CL) in Figure 4.
Note that the top scoring relationship found for DOID:10652 ("Alzheimer's disease") is a trivial one with HP:0002511 ("Alzheimer disease"), as this disease is also included in HPO's vocabulary. These trivial cases can be detected by the "string similarity" parameter (1.0 in this case, highlighted in red) and eventually discarded introducing a threshold in the text-box of that column (Figure 3).