[CSBG main page]
Biological significance of GO terms and relationships
"[..] the scientific community should recognize that classification is a purposeful human activity that reflects observations about relationships among properties of phenomena. As a variety of relationships occur in nature, different classification schemes can coexist. On the other hand, classification that does not reflect true relationships can misguide scientific discourse".
J. Parsons & Y. Wand "A question of class" Nature (2008) 455:1040-1041
"Categorizing is necessary for humans, but it becomes pathological when the category is seen as definitive, preventing people from considering the fuzziness of boundaries, let alone revising their categories."
N. N. Taleb. "The Black Swan - The Impact of the Highly Improbable".
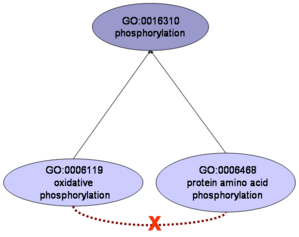
Classification is intrinsic to the scientific method. In many cases, the groups defined by a classification schema have a rationale behind, in the sense that elements grouped together share common properties. In other cases, the grouping is due to methodological or other reasons not related to the properties of the elements. Some authors term these two types of groups 'classes' and 'categories', respectively, depending on whether they group objects with common properties or not (Parsons and Wand, 2008). Categories are not necessarily bad, as long as they are recognized as such (mere classification schemes due to methodological needs) and not assumed to have the properties of classes.
The de-facto standard today for representing protein function, the Gene Ontology (GO), comprises a set of terms (vocabulary) related by parenthood relationships based on expert knowledge for describing different aspects of the complex phenomenon of "protein function". GO was created for the purpose of annotating proteins. Nevertheless, due to its relative simplicity and easiness to be handled by computers, it is increasingly used beyond that primary goal. The "biological process" subontology of GO (GO:BP) is widely used to evaluate sets of relationships between proteins (e.g. protein-protein interactions or co-regulation relationships) under the assumption that proteins annotated with the same (or related) GO:BP terms interact or are functionally related. This assumption only holds true for GO:BP terms with "biological significance" (classes) and not for the ones which are conceptualizations ("categories"). It is important to remark that the existence of "categories" is not a problem for the main goal of GO, protein annotation. Although the overall biological significance of the GO has been previously demonstrated, the functional coherence of individual GO terms and their relationships was not evaluated or quantified in an exhaustive way.
|
We addressed the evaluation of the significance of individual GO:BP terms using a high-quality functional network. We first distinguished the terms that are functionally coherent from those that are not ("classes" and "categories"). We also evaluated the relationships defined in the GO:BP hierarchy. Finally, we extracted those terms that are functionally unrelated according to GO, but which should be related according to the functional network.
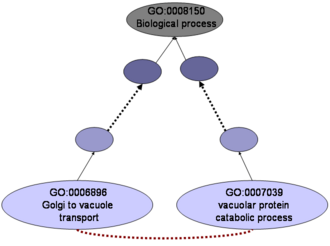
While many GO:BP terms are functionally coherent, we show examples of GO:BP terms not reflected in functional linkages between the proteins associated with them. These terms are intuitively recognized as categories. As expected, there is certain relationship between the specificity of the terms as extracted from the GO:BP hierarchy and functional coherence, although we have found cases of very specific terms which are not functionally coherent, and the other way around. Similarly, most parenthood relationships (which are explicit in GO:BP) are supported by functional data. But many brotherhood relationships (implicit in GO) are not. We also found new relationships between GO terms not apparent in the ontology (either explicitly or implicitly) and discuss their implication in the GO-based analysis of interaction data.
The main message of this work is that GO:BP terms and relationships are not equally supported by current functional associations. Therefore, they have to be used with caution when utilized for evaluating individual protein associations, a goal for which GO:BP was not specifically designed.
|
More information
Monica Chagoyen & Florencio Pazos. (2010).
Quantifying the biological significance of gene ontology biological processes - implications for the analysis of systems-wide data.
Bioinformatics.
26(3):378-384.
[PubMed:19965879]
[HTML]
[PDF]

